Introduction
Welcome to the Hemato.AI Diagnostic API! You can use our API to access Hemato.AI's Diagnostic AI and Clinical Algorithms, which processes images of peripheral blood slides and generates hematopathology reports.
Examples on this page are in shell script and language bindings in Go, Typescript, Python, C#, and TypeScript coming in future! You can view code examples in the dark area to the right, and you can switch the programming language of the examples with the tabs in the top right.
Endpoints
Hemato.AI API is made available at specific regional endpoints to adhere to regional regulations. Any data submitted to a regional endpoint will be handled on servers located within the same geographical and regulatory region.
All Hemato.AI API endpoints are only available through encrypted (TLS) connections.
There are a few special purpose endpoints that are not region specific: - For development and testing purposes anyone can use the Dev endpoint. To use this endpint you need to be assigned developer credentials. This endpoint is not HIPAA compliant and should not be used for any production purposes. Information submited to this service is not protected by any privacy laws and may be inspected by Hemato.AI staff. - For external quality assurance and external automated testing you can use the QA endpoint. Specially it is imporant to run automated benchmarks agains this endpoint before any significant change to your hardware or sofrwre that has the potential to affect the quality, or clinical validity of the outputs. To use this endpoint you need QA credentials. This endpoint is not HIPAA compliant and should not be used for any production purposes. Information submited to this service is not protected by any privacy laws and may be inspected by Hemato.AI staff.
You can explicitly choose what region to use from this list:
| Region | Endpoint | Users |
|---|---|---|
| DEV | https://dev.api.hemato.ai | for development and testing purposes |
| QA | https://qa.api.hemato.ai | for external quality assurance and external automated testing |
| USA | https://us.api.hemato.ai | Production, for customers in the United States |
| Canada | https://ca.api.hemato.ai | Production, for customers in Canada |
| EU | https://eu.api.hemato.ai | Production, for customers in the European Union |
HTTP Responses
Most Hemato.AI API will return a JSON encoded response in this format.
{
"status": 202,
"results": {
},
"error": "error message if any",
"user_message": "",
"debug_info": {
// helpful information for debugging, except for auth debugging, see Auth section below
}
}
| Field | Description |
|---|---|
| status | This reflects the HTTP Status code of the response. |
| error | If something has gone wrong, this provides an error message with error codes or more context on what has happened. |
| results | This is the output of the call, and can be of any type, often a dictionary / object. |
| user_message | This is a message that can be displayed to the end users. This is often useful in cases where there is an error. |
| debug_info | Contains helpful information for debugging |
Authentication
All authenticated calls to Hemato.AI API are expected to present an Authorization header containing a signed and valid token.
To obtain an authorization token you have 2 options:
- Session Based Authentication: you can call the
/auth/loginendpoint for a specific region and provide a user-name and password-hash (sha256 of the actual password) to obtain a newly minted and signed token. Session Auth is best suited for interactive applications where a user is directly involved in the process of obtaining the token. Example use cases are web applications, mobile apps, and desktop applications where individual users have Hemato.AI accounts. If you have a user facing application but your users don't hold individual Hemato.AI credentials, you can use the Public-Private Key authentication method below. - Public-Private Key: you can create your own Public-Private key paris, and register your public keys with Hemato.AI. Then you can sign your own tokens using the private key associate with the public keys you registered with Hemato.AI. This is particularly helpful in situations where a user is not directly involved and a device or an automated system needs to make calls to Hemato.AI API. Examples a hardware device digitizes samples and needs to call Hemato.AI to obtain an interpretation of the sample. Or you have a mobile app and allows the users to interact with Hemato.AI but the users don't hold individual Hemato.AI accounts. To learn more read How to register your public keys with hemato
Session Auth
# SECURITY RISK:
# Please ONLY use this for debugging login problems.
# MAKE SURE TO REMOVE THIS FROM YOUR SHELL HISTORY.
# otherwise your plan password will remain in your shell history and possibly leak into backups and such.
echo -n YOUR_ACTUAL_PASSWORD | shasum -a 256 | cut -d ' ' -f 1
package main
import (
"crypto/sha256"
"fmt"
)
func main() {
shasum := sha256.Sum256([]byte("YOUR_ACTUAL_PASSWORD"))
fmt.Printf("%x", shasum)
}
import { CryptoJS } from 'crypto'
const passSha = CtryptoJS.SHA256('YOUR_ACTUAL_PASSWORD').toString(CryptoJS.enc.Hex)
console.log(passSha)
For interactive use cases, where you or your users hold individual Hemato.AI accounts, you can use the Session Auth method. This method is best suited for web applications, mobile apps, and desktop applications where individual users have Hemato.AI accounts.
This call does NOT accept the raw user password. Instead you need to provide the sha256 hash of the password. This is to prevent the password from being leaked in internet trafic capture or intercepted on device or in transit. Of course all Hemato.AI endpoints are only available only through encrypted (TLS) connections. Still there are scenarios that a client browser or tools might allow for interception of traffic. You can use the following command to generate the sha256 hash of your password:
To authorize, using Session Authentication:
# SECURITY RISK:
# Please ONLY use this for debugging login problems.
# MAKE SURE TO REMOVE THIS FROM YOUR SHELL HISTORY.
# otherwise your plan password will remain in your shell history and possibly leak into backups and such.
export PASS_SHA_256=`echo -n YOUR_ACTUAL_PASSWORD | shasum -a 256 | cut -d ' ' -f 1`
# use httpie from https://httpie.io/
echo '{"user":"ali@example.com","pass_hash":"'${PASS_SHA_256}'"}' | http -F POST https://api.hemato.ai/auth/login
# or use curl, you can just pass the correct header with each request
echo '{"user":"ali@example.com","pass_hash":"'${PASS_SHA_256}'"}' | curl -X POST "https://api.hemato.ai/auth/login"
// Not implemented yet
The above command returns a JSON structure like this:
{
"status": 202,
"results": {
"login_timestamp": "1682945428455621477",
"token": {
"HY_APP_AUTH_v1": "eyJhbGciOiJIUzI1NiIsInR5cCI6IkpXVCJ9E....."
},
"user": {
"email": "ali@example.com",
"id": "03349841-588d-56b3-b93c-52d0983f9c75",
"screen_name": "Ali",
"user_name": "ali@example.com"
}
},
"debug_info": {
"delta": "463.021938ms",
"version": "bb_api.362.pbs_tasks.dfd119b"
}
}
Public / Private Key Auth
You can sign your own Authentications headers, see JWT specifications.
A self-service dashboard to register your public keys is coming soon. In the meantime, please contact us to register your public keys.
JWT Specifications
If you are using Public-Private Key Auth, you will be generating and signing your own Authorizaiton headers. Hemato.AI authorization headers are JSON Web Tokens (JWT) (more info at (https://jwt.io/)[https://jwt.io/]).
Suppoted Signing Algorithms
Curently only RS256 (RSASSA-PKCS-v1.5 using SHA-256) is allowed.
Required Fields
All the fields in this table are required. All keys and values are case sensitive. All JWTs need to be BASE64 encoded when used as Authorization headers.
| JWT Claim | Abbreviated | Data Type | Notes |
|---|---|---|---|
| Token ID | jti |
String | A unique ID for each token. Most useful for audit, debugging and investigation of issues. UUID or high resolution timestamps can be used. |
| Issuer | iss |
String | This is the identifier of your organization. |
| Issed at | iat |
Unix Time aka IntDate | The time the token was issues. A token issued in future is invalid. A token more than 1 hour is considered expired. |
| Expiraion | exp |
Unix Time aka IntDate | The time this token will expire and be invalid. A token more than 1 hour is considered expired, regardless of this value. |
| Audience | aud |
String | This needs to match the domain and region you are calling. Examples are: us.api.hemato.ai or dev.api.hemato.ai |
| User ID | sub |
String | This needs to be one of the user ids from the same organization where the signing key belongs to and this use must have permission to access this region. |
| Role | role |
String | This is the role of the user. Available roles are admin, user, device, service. This is used to determine the permissions of the bearer. |
| Key ID * | kid |
String | Only required if using RS256 signing algorithm. This is the ID of the public key that was used to sign the token. |
How to create your own keys
ssh-keygen -t rsa -b 4096 -m PEM -f jwtRS256.key
# Don't add passphrase
openssl rsa -in jwtRS256.key -pubout -outform PEM -out jwtRS256.key.pub
cat jwtRS256.key
cat jwtRS256.key.pub
Example
On the right you can see an example of a Hemato.AI JWT
This is the structure of a Hemato.AI JWT
json { "alg":"HS256", // or RS256 "typ":"JWT" } . { "aud": [ "us.api.hemato.ai", ], "exp": 1683095428, "iat": 1682945428, "iss": "1stdevision.example.com", "jti": "2d33d1d518", "sub": "01234567-789d-46b7-b38c-45d4562f5c12", "role": "user", "kid": "01234567-789d-46b7-b38c-45d4562f5c12" }
Debugging JWTs
You can decode and inspect your JWTs at https://jwt.io/
You can also make a call to /auth/hello endpoint and get more helpful information about your JWT and reasons it may be rejected.
# if you are using httpie
http -f POST https://dev.api.hemato.ai/auth/helo Authorization:HEMATO_AI_AUTH_TOKEN
# if you are using curl
curl -x POST --header "Authorization:HEMATO_AI_AUTH_TOKEN" https://api.hemato.ai/auth/hello
Get a PBS Report
Images of Peripheral Blood Smears (PBS) usually are very large. It is often impractical to submit an entire sample in one API call. That's why there are a few steps to submit a PBS for review by Hemato.AI.
There are two workflows available to get Hemato.AI to study a peripheral blood sample and generate morphological, diagnostic and pathology reports.
In Workflow 1, you make API calls to upload the images, request a diagnostic study and get the results.
In Workflow 2, you make an API call to provide a link where Hemato.AI can download your files and then you can make an API call to get the results.
Workflow 1
In workflow 1, to get Hemato.AI's opinion on a Peripheral Blood Smear you follow these steps:
- Get an ID for your new PBS
- Upload the files under the new ID
- Request a Diagnostic Study
- Wait for it
- Ask for the report
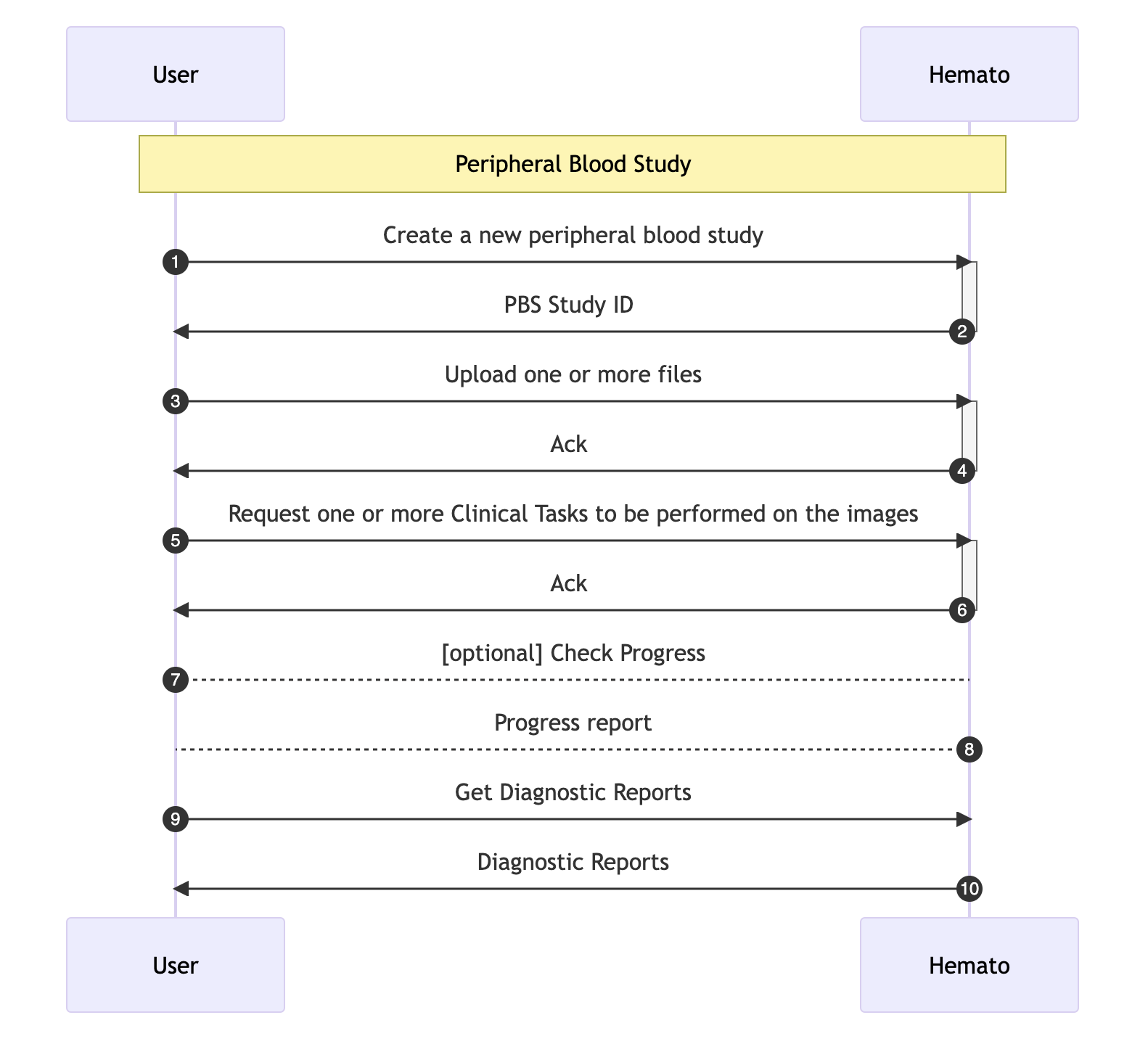
1. Get an ID
Get a new PBS ID. This ID will be used to upload the images, request diagnostic study of the sample, provide any other information available and afterwards get the results of Hemato.AI's review.
HTTP Request
Make a POST call to the /pbs endpoint. You can provide any number of tags (key value string pairs) along side this request. These tags can allow you to associate Patient Proxy Identifiers or Organization IDs or any number of other information that is important to you with the PBS. These tags can later be used to find and retrieve PBSs easier.
# use httpie from https://httpie.io/
echo '{"tags":{"patient_proxy_id":""}}' | http -f POST https://api.hemato.ai/pbs Authorization:HEMATO_AI_AUTH_TOKEN
# alternatively use curl
curl -x POST --header "Authorization:HEMATO_AI_AUTH_TOKEN" https://api.hemato.ai/pbs
Make sure to replace
HEMATO_AI_AUTH_TOKENwith your API key.The above command returns a JSON structure like this:
{
"status": 201,
"results":{
"pbs_study_id":"c7d453a9-676d-4b9f-a505-8a9021b76dfd"
}
}
2. Upload the files
Upload the sample by calling the PBS upload API as many times as needed.
HTTP Request
Make a POST call to the /pbs/YOUR_NEW_PBS_ID/files?file_name=some_file_name.jpg endpoint. Here YOUR_NEW_PBS_ID is the id you obtained from step 1 (results.pbs_id in the response structure returned).
To determin the file type, this endpoint expects either "file_name" or "content_type" query parameters. For example for a JPEG file you can use file_name=some_file_name.jpg or content_type=image/jpeg.
This is an idempotent call. It means you can send the same file multiple times and it will only be counted as one file.
Once submitted however, you cannot update the same file or change it or its metadata. You can only add more files to the same PBS study.
The file type is determined by the `Content-Type` header of your request. So when sending a request to upload a jpg file for example, your request needs to have a header `Content-Type:image/jpeg`.
This endpoint accepts:
File Type | Request Content-Type Header
--------- | -----------
jpeg , jpg | image/jpeg
png | image/png
zip ★ | application/zip
★ Zip files can contain jpg or png files.
If the Content-Type header is missing, the file name extension for "file_name" provided is used to determine the file type.
If no file type can be determined or provided, a 400 - Bad Request error is returned.
```shell
# use httpie from https://httpie.io/
http -f POST https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/files Authorization:HEMATO_AI_AUTH_TOKEN Content-Type:image/jpeg < /path/to/filename.jpg
# alternatively use curl
curl -x POST --header "Authorization:HEMATO_AI_AUTH_TOKEN" --header "Content-Type:image/jpeg" --data-binary "@/path/to/filename.jpg" https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/files
The above command returns a JSON structure like this:
{
"status": 201,
"results":{
"pbs_study_id":"c7d453a9-676d-4b9f-a505-8a9021b76dfd",
"file_id":"sha224-i-4d58b0cee1e38f75f93e2e605d1e6ad44c24a975d0c3b53367de7c86"
},
"debug_info":{
}
}
3. Request a Diagnostic Study
When all the files for a particular PBS is uploaded, you will make a call to mark the PBS as ready to be processed and ask for any number of diagnostics tasks to be performed on this PBS. After this call you will not be able to upload additional files for this PBS.
Optionally you can register a callback_url that will be called when the report is ready.
We’ll send a POST request to the callback URL with details of the report when ready.
The callback url needs to accept a POST call, and must support HTTPS with a valid server certificate. The body of the call will conform to {"pbs_stody_id":"25f7de38-ba9c-4b45-a4d8-8c13461d7b39", "report_status": "ready", "progress": 1.0} but it can be ignored.
So there are two approaches that users can take:
1. Include the pbs_study_id or any other information you need (no PII or PHI) in the url to be called.
2. Use a single generic url, but parse and use the information in the body to identify which study is the subject of the call back.
Although it is not strictly needed, we recommend you include a cryptographic signature as part of your URL, to that you can verify the authenticity of the cells to your endpoint. Example:
{"callback_url":"https://example.com/pbs_report_is_ready/<PBS_STUDY_ID>/<timestamped_cryptographic_signature_valid_only_for_this_PBS_STUDY_ID>"
This enables early filtration of incoming traffic, particularly in cases where the backend systems may be inundated by a surge of malicious or rogue requests.
# use httpie from https://httpie.io/
echo '{"diagnostic_tasks":["pbs_v1"], "callback_url":"https://example.com/pbs_report_is_ready/<PBS_STUDY_ID>/"}' | http -f POST https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/tasks Authorization:HEMATO_AI_AUTH_TOKEN
# alternatively use curl
echo '{"diagnostic_tasks":["pbs_v1"], "callback_url":"https://example.com/pbs_report_is_ready/<PBS_STUDY_ID>/"}' | curl -x POST --header "Authorization:HEMATO_AI_AUTH_TOKEN" --data-binary @- https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/tasks
The above command returns a JSON structure like this:
{
"results":{
"pbs_study_id":"48ae6352-d15d-440a-83f3-05a1f68aa11b",
"tasks": [
"pbs_v1"
]
}
}
4. Wait for it
Depending on the size of the files submitted, the number Diagnostic tasks requested, and other factors like general system workload, it will take between a few seconds to 10s of seconds for the results to be ready.
If you registerd a callback url in step 3, you can wait for that url to be called.
You can check the status of the task by making a GET call to /status
To get the status of a PBS
#
http https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/status Authorization:HEMATO_AI_AUTH_TOKEN
#
curl --header "Authorization:HEMATO_AI_AUTH_TOKEN" https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/status
This returns
json { "status": 200, "results": { "pbs_study_id": "62dd0300-880e-4069-bea8-66c9ca73c207", "report_status": "ready", "progress": 1.0, "progress_message":"Reports are ready", }, "debug_info" :{} }
5. Get The Report for a Peripheral Blood Smear Study
You can get the latest version of the report by making a GET call. In cases that there are multiple revisions and you want to get an earlier version of the report you need to specify the version.
To get the latest diagnostic report on a PBS
#
http https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/report Authorization:HEMATO_AI_AUTH_TOKEN
#
curl --header "Authorization:HEMATO_AI_AUTH_TOKEN" https://api.hemato.ai/pbs/YOUR_NEW_PBS_ID/report
Which in turn returns a JSON structure like:
{
"status": 200,
"results": {
"pbs_study_id": "4bb7fe9e-b608-4d68-adbe-8655c991f494",
"file_ids": ["sha224-i-db367ccd89fc39aa6ff9fff0cd9b11e6b5b6a41cff4bbb232bee7c93"],
"feature_statistics": {
"rbc-density": {
"value": "x",
"unit": "y"
},
"platelet-density": {
"value": "x",
"unit": "y"
},
"blast-density": {
"value": "x",
"unit": "y"
}
},
"morphological_findings": {
"ring trophozoite": {
"count": "1",
"heat_map": [{
"center": [435, 345],
"radius": 35
}]
}
},
"diagnostic_report": {
"differential_diagnosis": [{
"score": 7,
"confidence": "0.95",
"clinical-significance": "D",
"icd10-code": "B50",
"english-long": "Malaria Infection with Plasmodium Falciparum",
"english-short": "Malaria"
}]
},
"pathology_report": "No demographics or past medical history is available about the patient. Inspection of peripheral blood slide revealed ring trophozoites ..."
},
"debug_info": {}
}
There are 3 levels of report available.
- Morphological Findings
- Diagnostic Information
- Pathology Report
Workflow 2
In workflow 2, to get Hemato.AI's opinion on a Peripheral Blood Smear you follow these steps:
- Submit a url from which Hemato.AI can download the files and get an Study ID back (you can use this ID to check the status of the study and get the report)
- Optionally provide a callback url
- Wait for it
- Get a callback, with information about the sample, dwnloaded file size and formats and sanity checks.
- Wait for it
- Get a callback, with information about the report, and a link to the report
- Check the status of the study and get the report
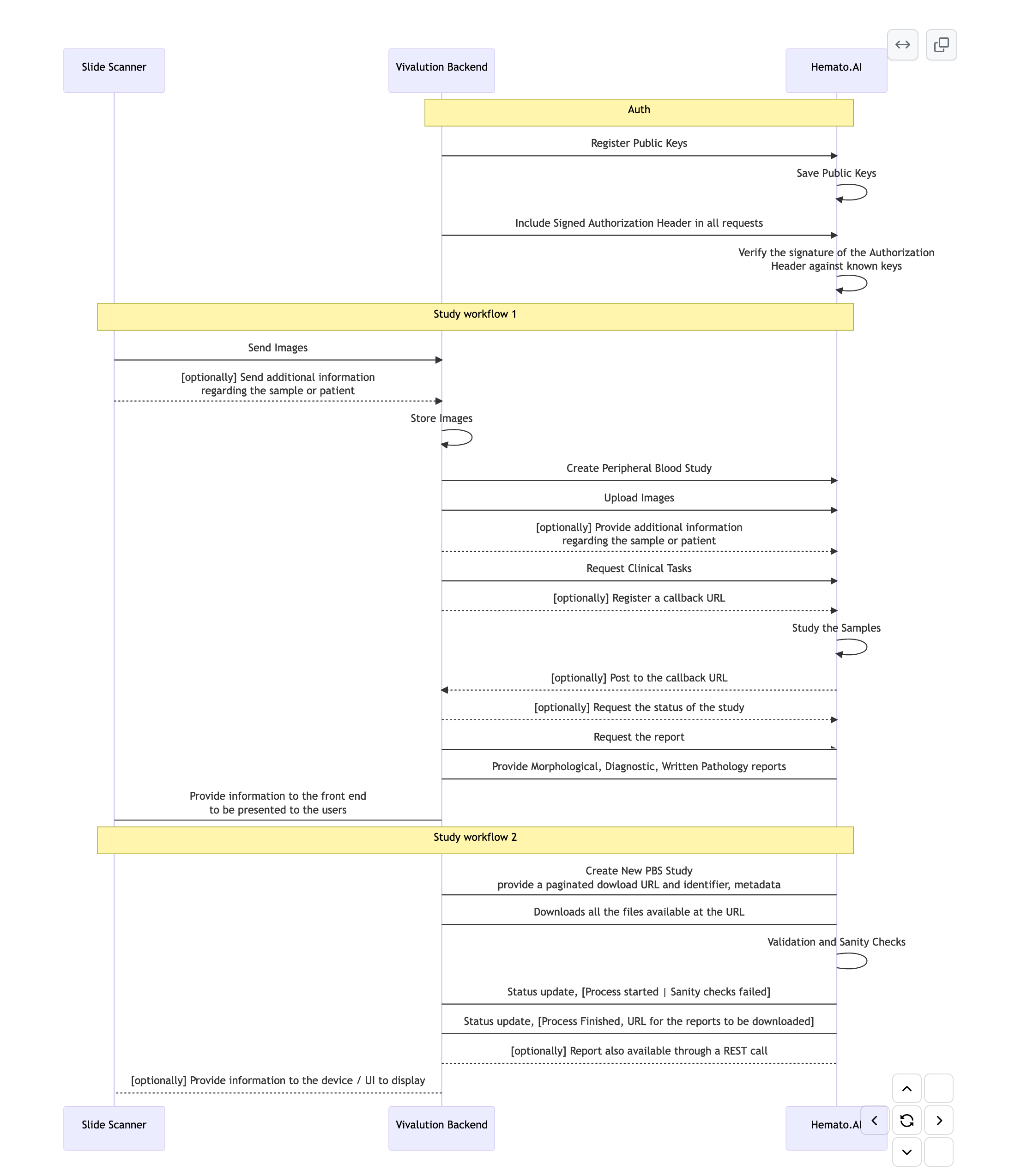
1,2,3. Submit a url from which Hemato.AI can download the files and get an Study ID back
You can make a POST call to /pbs with a url to retrieve a list of files from. this call will return a Study ID that you can use to check the status of the study and get the report.
The call needs to include a list of diagnstic tasks that you want to be performed on the files.
The list of files returned by the provided list_url can be a list of presigned urls to files in an S3 bucket, or a list of urls to files in a publically accessible location, or in some other way need to be accessible to Hemato.AI.
The body of this POST request will look like the example on the right:
Request body to start "Workflow 2"
{
"list_url": "https://example.com/samples/1/files.json",
"tasks": [
"pbs_v1"
]
}
On the right you can see an example of the list of files that will be downloaded by Hemato.AI as a single periheral blood study.
{
"peripheral_blood_sample_id": "62dd0300-880e-4069-bea8-66c9ca73c207",
"patient_proxy_medical_record_number": "123456789",
"sample_collection_timestamp": "2021-01-02T15:0405+07:00", // RFC3339 formated date time
"clinical_tasks_requested": [
"pbs_v1"
],
"files": [
"https://presignedurldemo.s3.eu-west-2.amazonaws.com/image.png?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAJJWZ7B6WCRGMKFGQ%2F20180210%2Feu-west-2%2Fs3%2Faws4_request&X-Amz-Date=20180210T171315Z&X-Amz-Expires=1800&X-Amz-Signature=12b74b0788aa036bc7c3d03b3f20c61f1f91cc9ad8873e3314255dc479a25351&X-Amz-SignedHeaders=host",
...
],
"callback_url": "https://example.com/callbacks/1"
}
To submit a url to a Peripheral Blood Smear Study
echo '{"peripheral_blood_sample_id":"xyz", "patient_proxy_medical_record_number":"abc", ... }' | http -F POST https://dev.api.hemato.ai/pbs Authorization:HEMATO_AI_AUTH_TOKEN
{
"results": {
"pbs_study_id": "4bb7fe9e-b608-4d68-adbe-8655c991f494",
"file_ids": ["sha224-i-db367ccd89fc39aa6ff9fff0cd9b11e6b5b6a41cff4bbb232bee7c93"]
},
}
4,5 Receive a callback with information about the sample, downloaded file size and formats and sanity checks.
When the study has been downloaded and the files have been checked for sanity, you will receive a callback to the callback_url you provided in the previous step.
The example on the right shows the body of a callback request.
{
}
6 Receive a callback with information about the report, and a link to the report
When the study has been analysed and the report is ready, you will receive a callback to the callback_url you provided in the previous step.
The example on the right shows the body of a callback request.
{
}
7 Check the status of the study and get the report
You can make a GET call to /pbs/{pbs_study_id}/status to get the status of the study and the report.
To retrive the reports you can make a call to /pbs/{pbs_study_id}/report.
for more information see the same call in "workflow 1" above (step: get the report for peripheral blood study).
Health Check
If you need to check the health status of the Hemato.AI api you can make a GET call to the heartbeat endpoint.
http https://api.hemato.ai/heartbeat
{
"delta": "59.078µs",
"heartbeat": "a7ef8fba741237f693c3",
"request_timestamp": 1680716829,
"version": "bb_api.331.develop.18ff645"
}
Errors
The Hemato.AI API uses the following error codes:
| Error Code | Meaning |
|---|---|
| 400 | Bad Request -- Your request is invalid. |
| 401 | Unauthorized -- Your API key is not correctly signed or expired. |
| 403 | Forbidden -- Your token does not grant you permission to perform this task. |
| 404 | Not Found -- The specified resource could not be found. |
| 405 | Method Not Allowed -- You tried to access a resource with an invalid method. |
| 406 | Not Acceptable -- You requested a format that isn't json. |
| 418 | I'm a teapot. (for debugging purposes) |
| 429 | Too Many Requests -- You're being throttled |
| 500 | Internal Server Error -- Our apologies, We had a problem with our server. Please try again later. |
| 503 | Service Unavailable -- Our apologies, We're temporarily offline. Please try again later. |

